I have decided to write a tutorial on a topic not even Um_nik knows! (source)
In this tutorial, I will talk about the blossom algorithm, which solves the problem of general matching. In this problem, you are given an undirected graph and you need to select a subset of edges (called matched edges) so that no two edges share a vertex, and the number of matched edges is maximized. More common in competitive programming is bipartite matching, which is the same problem but when the graph is guaranteed to be bipartite. In competitive programming, general matching seems to get a lot of hate for being very challenging to implement. However, the blossom algorithm is quite beautiful, and important in the history of algorithm research.
It will help if you are already familiar with bipartite matching. I will discuss the high level ideas of the algorithm, but my main focus will be on the tricky implementation details. So you may also want to read a supplementary resource like the Wikipedia page (from which I stole some pretty pictures).
Algorithm Overview
The blossom algorithm works by increasing the number of matched edges by one at a time until it cannot increase it further, then it stops. It's not as simple as just adding a new edge and keeping all the edges from previous iterations. If we did this, we might make a wrong decision and have a sub-optimal matching. Instead of simply searching for one new edge to add to the set, we should search for an augmenting path. This is a path where the edges alternate between being matched and unmatched. A vertex is called exposed if it is not adjacent to any matched edge. Another requirement of augmenting paths is that the endpoints should both be exposed. Also, it must be a simple path, meaning it cannot repeat any vertices.
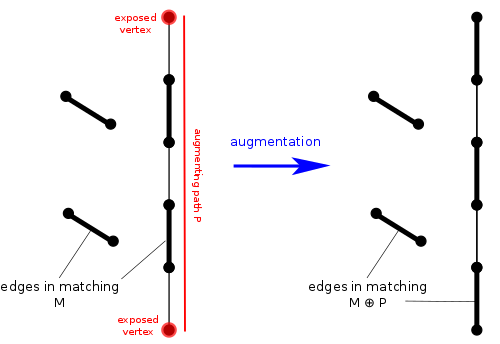
If we find an augmenting path, we can easily increase the number of matched edges by $$$1$$$. We simply change all the matched edges to unmatched and all the unmatched edges to matched. An augmenting path has one more unmatched edge than matched edges, so it's correct. We can also easily see that it will still be a valid matching, i.e. no two matched edges will share a vertex. Ok, so augmenting paths are useful for increasing our matching, but are they guaranteed to exist if we don't have a maximum matching? Yes! In fact, consider $$$M_1$$$ to be the set of edges in our current matching and $$$M_2$$$ to be the set of edges in some maximum matching. Now, the symmetric difference $$$M_1\oplus M_2$$$ will be all the edges in exactly one of the two matchings. Based on the matching property, each vertex has degree $$$1$$$ or $$$2$$$. So each connected component must be a cycle or a path. Since we assume $$$M_2$$$ has more edges than $$$M_1$$$, there must be some connected component in $$$M_1\oplus M_2$$$ with more edges from $$$M_2$$$ than $$$M_1$$$. The only way this can happen is if one of the components is an augmenting path.
Great! Now all we have to do is figure out how to find an augmenting path in a matching, and we're done. Unfortunately, this is more difficult than it sounds. We might think of just using a DFS or BFS from exposed vertices, and ensuring that edges always alternate between matched and unmatched. For example, we could say each vertex $$$v$$$ has two versions $$$(v, 0)$$$ and $$$(v, 1)$$$. We can ensure that matched edges always go from version $$$1$$$ to $$$0$$$ and unmatched from $$$0$$$ to $$$1$$$. So, what's wrong? Won't we correctly find an alternating path of matched and unmatched edges, from one exposed vertex to another? The issue is that we are not ensuring the path is simple. This process could find a path that visits both $$$(v, 0)$$$ and $$$(v, 1)$$$ for some $$$v$$$. Then if you "flip" all the edges, $$$v$$$ would be adjacent to two matched edges, which is a violation. However, this simple DFS/BFS idea does work correctly in bipartite matching, and you might see it's equivalent to the algorithm you already know.
Blossoms
To deal with the problem of repeating vertices, we introduce the concept of blossoms. A blossom is simply a cycle of $$$2k+1$$$ edges, where $$$k$$$ of them are matched edges. This means there is exactly one "special" vertex $$$v$$$ in the cycle that isn't matched to another vertex in the cycle. Why do we care about blossoms? Because they create a situation where an alternating DFS/BFS could repeat a vertex.
Now that we know what they are, how does the blossom algorithm handle them? It contracts them. This means that we merge all vertices of the blossom into a single vertex, and continue searching for an augmenting path in the new graph.
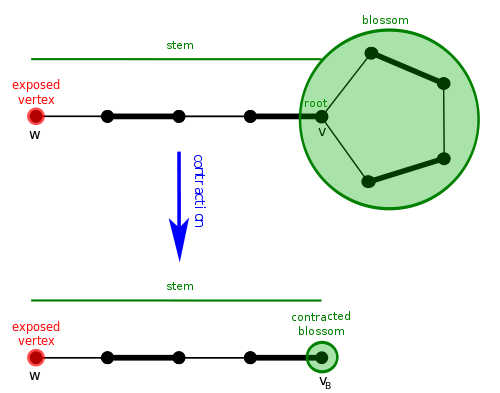
If we eventually find an augmenting path, we will have to lift the path back to our original graph. It can be shown that a graph has an augmenting path if and only if it has one after contracting a blossom. The concept of contracting blossoms and lifting an augmenting path makes it challenging to implement correctly. Remember, a contracted blossom can become a vertex in another blossom, so you can have blossoms inside blossoms inside blossoms! You should be experiencing headaches around now.
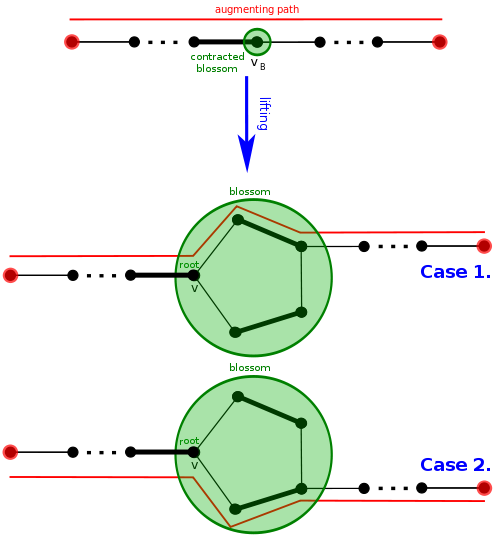
Let's see intuitively how an augmenting path can be lifted, effectively "undoing" a blossom contraction while keeping an augmenting path. Well, there should be an edge entering the blossom and then an edge leaving it. Since the path is alternating in the contracted graph, one of those two edges is matched. This means the "special" vertex $$$v$$$ of the blossom will be involved. Suppose $$$u$$$ is the vertex involved with the unmatched edge leaving the blossom. Let's go around the cycle between $$$u$$$ and $$$v$$$, but there are two ways, which do we take? Of course, it should be alternating, so we have exactly one correct choice.
Summary
In summary, the algorithm works like this. We repeat the following process until we fail to find an augmenting path, then return. We begin a graph search with DFS or BFS from the exposed vertices, ensuring that the paths alternate between matched and unmatched edges. If we see an edge to an unvisited node, we add it to our search forest. Otherwise if it's a visited node, there are three cases.
- The edge creates an odd cycle in the search tree. Here, we contract the blossom and continue our search.
- The edge connects two different search trees and forms an augmenting path. Here, we keep undoing the blossom contractions, lifting the augmenting path back to our original graph, and flip all the matched and unmatched edges.
- The edge creates neither case 1 nor case 2. Here, we do nothing and continue our search.
Implementation in $$$O(n^3)$$$
For the implementation, I will use an integer ID for each vertex and for each blossom. The vertices will be numbered from $$$0$$$ to $$$n-1$$$, and blossoms will start at $$$n$$$. Every blossom contraction gets rid of at least $$$3$$$ previous vertices/blossoms, so there can be at most $$$m:=\frac{3n}{2}$$$ IDs. Now, here is my organization of all the data:
mate: an array of length $$$n$$$. For each vertexu, if it's exposed we havemate[u] = -1, otherwise it will be the vertex matched tou.b: for each blossomu,b[u]will be a list of all the vertices/blossoms that were contracted to formu. They will be listed in cyclic order, where the first vertex/blossom in the list will be the "special" one with an outgoing matched edge.bl: an array of length $$$m$$$. For each vertex/blossomu,bl[u]will be the blossom immediately containingu. Ifuis not contracted inside of another blossom, thenbl[u] = u.p: an array of length $$$m$$$. For each vertex/blossomu,p[u]will be the parent vertex/blossom ofuin the search forest. However, we will be a bit relaxed: we also allow it ifp[u]is contracted inside the real parent, or even contracted multiple times, as long as the vertex/blossom at the top is the real parent in the contracted graph.d: an array of length $$$m$$$. For each vertex/blossomu,d[u]will be a label/mark telling its status in the search forest. We will assignd[u] = 0if it's unvisited,d[u] = 1if it's an even depth from the root, andd[u] = 2if it's an odd depth from the root.g: a table of size $$$m\times m$$$, storing information about the unmatched edges. For each pair of vertices/blossoms $$$(u, v)$$$, theng[u][v] = -1if there is no unmatched edge between them. Otherwise if there's an unmatched edge, then we will use this table entry to help us with lifting augmenting paths. When we're lifting a path through a blossom, we would like to know which vertices inside the blossom need to be connected. So ifuis a blossom, theng[u][v]will store the vertex inside the blossom ofuthat connects tov. Otherwise ifuis a vertex, theng[u][v] = u.
Structure
Now, we can define the structure and a couple helper functions add_edge and match. We use add_edge to create an unmatched edge, and match to change an unmatched edge to matched.
Trace Path to Root
We will want a function that traces the path to the root, where we only take vertices/blossoms in the contracted graph. This is done by repeatedly finding the blossom at the top of the bl chain, and following the parent pointers p.
The complexity of this function is $$$O(n)$$$.
Blossom Contraction
Let's say we found an edge between vertices $$$x$$$ and $$$y$$$ that creates a blossom in the search forest, and we need to contract it. Let's say that $$$c$$$ should be the ID of the new blossom, and we've constructed the paths from $$$x$$$ and $$$y$$$ to the root (call the paths vx and vy). First, we need to find the special vertex of the blossom, which is given by the lowest common ancestor of $$$x$$$ and $$$y$$$. So, we can say $$$r$$$ is the last common element of the vectors vx and vy, and delete everything above and including $$$r$$$.
Next, we should define b[c] to be the blossom vertices in cyclic order, starting at $$$r$$$. Simply append vx in reverse order, then vy in forward order. Finally, we should make the g table correct for the blossom c. Simply look at each vertex z in the blossom and each edge of z.
The complexity of this function is $$$O(n|b_c|)$$$ if the number of vertices/blossoms in $$$c$$$ is $$$|b_c|$$$.
Path Lifting
Let's say that we have an augmenting path in the contracted graph, and we want to lift it back to the original graph. The input will be a list of blossoms, where each one connects to the next, and we want to expand all of the blossoms except the last one, and return the list A of vertices.
The input list will work like a stack. If the top is a vertex, we will add it to the output and continue. Otherwise, we will replace the top blossom with the path of blossoms/vertices inside it such that it's still an alternating path. The variables represent the following information:
z: the top of the stackw: the next element on the stack afterzi: the index in theb[z]list of the last vertex on our lifted pathj: the index in theb[z]list of the first vertex on our lifted pathdif: the direction we should advanceiuntiljso that the path is correctly alternating.
As you can see, we use the g table to find the vertices/blossoms at the level below z. We also use the parity of the size of A to determine if the incoming edge is matched or unmatched.
The complexity of this function is $$$O(n)$$$.
Putting it all Together
Now that we have the subroutines, let's implement the whole algorithm. First, the algorithm will repeatedly search for augmenting paths until we can't find one and return. So we have one big outer loop to count the number of edges in our matching. Next, to start an iteration we reset all our variables, and assume the g table is correct for the vertices. We will use a BFS-like process for the search forest, but something like DFS should also work. We look for all the exposed vertices and add them to the queue. The variable c will be used to count the total number of vertex/blossom objects, and we'll increment it with each blossom contraction.
When we dequeue a vertex $$$x$$$, assuming it's not contained in another blossom, we look at all unmatched edges leaving it. Say we're looking at an edge to another vertex $$$y$$$. There are several cases:
- The vertex $$$y$$$ is not visited. In this case, we will mark $$$y$$$ and its mate as visited, and add
mate[y]to the queue. - The vertex $$$y$$$ is visited and has an odd distance to the root. In this case, we should do nothing.
- The vertex $$$y$$$ is visited and has an even distance to the root. Here, we should trace the paths from $$$x$$$ and $$$y$$$ to the root to determine if this event is a blossom contraction or an augmenting path. In either case, we should break from the inner loop.
Let's analyze the time complexity. First, each iteration increases the number of matched edges by $$$1$$$ and so there can be at most $$$n/2$$$ iterations overall. The basic BFS operations will clearly take $$$O(n^2)$$$ time since each vertex appears in the queue at most once, and each dequeued element looks at all other vertices. An augmenting path is found at most once in an iteration, and it takes $$$O(n)$$$ time. A blossom contraction takes $$$O(n |b_c|)$$$ time, and each vertex/blossom will belong to at most one immediate blossom, so the overall time in an iteration from blossom contractions is $$$O(n^2)$$$. Since each iteration takes $$$O(n^2)$$$ time and there are $$$O(n)$$$ iterations overall, we see that the algorithm takes $$$O(n^3)$$$ time.
I've tested my implementation on these two sites. If you want to do further testing/benchmarking, feel free, and let me know if you find any issues.










I'm more interested in how to blossom like Um_nik, without knowing such algorithms.
IIRC there's a randomized algorithm with better asymptotics (but worse constant factors). Does anyone have a submission implementing it so we can compare?
Here’s a previous discussion about General Graph Matching. https://mirror.codeforces.com/blog/entry/63630
Here is a comment with a link to a randomized algorithm submission. https://mirror.codeforces.com/blog/entry/63630?#comment-475059
I’m not sure if this is the one you’re thinking of.
I was thinking of this paper https://www.mimuw.edu.pl/~mucha/pub/mucha_sankowski_focs04.pdf which was linked to by the wiki article.
The blog post you linked still had a lot of useful info/problems/implementations so thanks for that!
The part about faster asymptotics is mostly theoretical, since for realistic sized matrices it's still best to do $$$O(n^3)$$$ matrix multiplication.
But still, it's good to be aware of the Tutte matrix and its application to maximum matching. You can use it to find the size of the maximum matching a lot easier than constructing it, for example.
Oh, nice, e-maxx only has $$$O(n^4)$$$ approach by Rabin and Vazirani with Tutte's matrix. This paper actually shows how to improve it further to $$$O(n^3)$$$ and beyond. And in a very reasonable way I must say.
Ah, I forgot that was randomizes.
Here’s an implementation of that paper. https://github.com/kth-competitive-programming/kactl/blob/main/content/graph/GeneralMatching.h
I initially wanted to write a separate blog post about it, but I just can't do that without properly explaining why $$$\text{Pf }^2A = \det A$$$ (which I can't do at the moment because I don't see meaningful explanation for it and proofs I find elsewhere are crazy), so I just post a brief summary of that randomization algorithm here.
The Tutte matrix of the graph $$$G=(V, E)$$$ is
\begin{equation} A(x_{12}, \dots, x_{(n-1)n}) = \begin{pmatrix} 0 & x_{12} e_{12} & x_{13} e_{13} & \dots & x_{1n} e_{1n} \newline -x_{12} e_{12} & 0 & x_{23} e_{23} & \dots & x_{2n} e_{2n} \newline -x_{13} e_{13} & -x_{23} e_{23} & 0 & \dots & x_{3n} e_{3n} \newline \vdots & \vdots & \vdots & \ddots & \vdots \newline -x_{1n} e_{1n} & -x_{2n} e_{2n} & -x_{3n} e_{3n} & \dots & 0 \end{pmatrix} \end{equation}
Here $$$e_{ij}=1$$$ if $$$(i,j)\in E$$$ and $$$e_{ij}=0$$$ otherwise, $$$x_{ij}$$$ are formal variables. Key facts:
Randomization comes when we substitute $$$x_{ij}$$$ with random values. It can be proven that conditions above still hold with high probability (usually $$$\frac{n}{|S|}$$$ is mentioned from Schwartz–Zippel lemma where $$$S$$$ is the set $$$x_{ij}$$$ come from). This provides us with $$$O(n^3)$$$ Gaussian elimination algorithm to find maximum cardinality matching in general graph.
UPD: Specifically for Tutte's matrix I see that there is some explanation to first property without involving Pfaffians, so I might write something about it after all.
tldr
Great blog! If you want a compact implementation in $$$O(nm)$$$ (with a little bit less bookkeeping than your approach), I've added it below. It should be suitable for things like ICPC notebooks.
Can you actually show that this is $$$O(NM)$$$? I've been using a similar implementation for a while but I recently realized that I don't actually know why it works (even in $$$O(N^3)$$$), considering that orig[x] does not actually store the current blossom x is contained in. Specifically, if there is some w such that orig[w] = orig[v] before the line "blossom(x, v, a); blossom(v, x, a);" runs, it is possible that orig[w] != orig[v] after that line runs.
Hope this comment explains the $$$O(N^3)$$$ run time!
Thanks for the nice blog! If anyone is interested, there is also an $$$O(m \sqrt{n} \log_{\max(2, 1 + m/n)} n)$$$ time and $$$O(n + m)$$$ space algorithm for general matching based on this paper, which also uses blossoms. The paper claims that it is possible to do it in $$$O(m \sqrt{n})$$$, but I use an implementation that has the previously mentioned complexity since it's easier to implement (basically lesser lines of code).
The implementation I use is here, but I forgot about the source of the implementation (it'd be great if someone could point to a source with the original implementation).
This implemention is by Min_25 and can be found here. The log factor just stems from the use of union find.
Min_25 also implemented an $$$O(n m \log(n))$$$ algorithm for max weight general matching here.
thanks a lot Grand Master Gon amazing insights received
Why does this hold in this case:
We have a cycle of 5 vertices, and 5 vertices, each of which is connected with an edge to a distinct vertex of the cycle.
Our current matching has 2 edges of the cycle, and an edge from the remaining vertex of the cycle to its pair.
There exists a perfect matching. cycle is a blossom. However, if we contract it, there is no augmenting path (what is left is a star graph with one edge already matched.
For the cycle to be a blossom, the "stem" (the matched edge hanging off the cycle) must be reachable from a free node. The algorithm does fine in this case. It does the DFS from one of the free nodes, and actually doesn't contract the cycle, but just finds an augmenting path between two of the vertices not in the cycle.
thanks, I missed that that was required for a blossom.